The @pewresearch folks have been collecting political survey data for quite a while, and I noticed the [visualization below](http://www.people-press.org/2014/06/12/section-1-growing-ideological-consistency/#interactive) referenced in a [Tableau vis contest entry](https://www.interworks.com/blog/rrouse/2016/06/24/politics-viz-contest-plotting-political-polarization):

Those are filled [frequency polygons](http://onlinestatbook.com/2/graphing_distributions/freq_poly.html), which are super-easy to replicate in ggplot2, especially since Pew even _kind of_ made the data available via their interactive visualization (it’s available in other Pew resources, just not as compact). So, we can look at all 5 study years for both the general population and politically active respondents with `ggplot2` facets, incorporating the use of `V8`, `dplyr`, `tidyr`, `purrr` and some R spatial functions along the way.
The first code block has the “data”, data transformations and initial plot code. The “data” is really javascript blocks picked up from the `view-source:` of the interactive visualization. We use the `V8` package to get this data then bend it to our will for visuals.
library(V8)
library(dplyr)
library(tidyr)
library(purrr)
library(ggplot2) # devtools::install_github("hadley/ggplot2)
library(hrbrmisc) # devtools::install_github("hrbrmstr/hrbrmisc)
library(rgeos)
library(sp)
ctx <- v8()
ctx$eval("
var party_data = [
[{
name: 'Dem',
data: [0.57,1.60,1.89,3.49,3.96,6.56,7.23,8.54,9.10,9.45,9.30,9.15,7.74,6.80,4.66,4.32,2.14,1.95,0.87,0.57,0.12]
},{
name: 'REP',
data: [0.03,0.22,0.28,1.49,1.66,2.77,3.26,4.98,5.36,7.28,7.72,8.16,8.86,8.88,8.64,8.00,6.20,5.80,4.87,4.20,1.34]
}],
[{
name: 'Dem',
data: [1.22,2.78,3.28,5.12,6.15,7.77,8.24,9.35,9.73,9.19,8.83,8.47,5.98,5.17,3.62,2.87,1.06,0.75,0.20,0.15,0.04]
}, {
name: 'REP',
data: [0.23,0.49,0.65,2.23,2.62,4.06,5.02,7.53,7.70,7.28,7.72,8.15,8.87,8.47,7.08,6.27,4.29,3.99,3.54,2.79,1.03]
}],
[{
name: 'Dem',
data: [2.07,3.57,4.21,6.74,7.95,8.41,8.58,9.07,8.98,8.46,8.47,8.49,5.39,3.62,2.11,1.98,1.00,0.55,0.17,0.17,0.00]
}, {
name: 'REP',
data: [0.19,0.71,1.04,2.17,2.07,3.65,4.92,7.28,8.26,9.64,9.59,9.55,7.91,7.74,6.84,6.01,4.37,3.46,2.09,1.65,0.86]
}],
[{
name: 'Dem',
data: [2.97,4.09,4.28,6.65,7.90,8.37,8.16,8.74,8.61,8.15,7.74,7.32,4.88,4.82,2.79,2.07,0.96,0.78,0.41,0.29,0.02]
}, {
name: 'REP',
data: [0.04,0.21,0.28,0.88,1.29,2.64,3.08,4.92,5.84,6.65,6.79,6.92,8.50,8.61,8.05,8.00,7.52,7.51,5.61,4.17,2.50]
}],
[{
name: 'Dem',
data: [4.81,6.04,6.57,7.67,7.84,8.09,8.24,8.91,8.60,6.92,6.69,6.47,4.22,3.85,1.97,1.69,0.66,0.49,0.14,0.10,0.03]
}, {
name: 'REP',
data: [0.11,0.36,0.49,1.23,1.35,2.35,2.83,4.63,5.09,6.12,6.27,6.41,7.88,8.03,7.58,8.26,8.12,7.29,6.38,5.89,3.34]
}],
];
var party_engaged_data = [
[{
name: 'Dem',
data: [0.88,2.19,2.61,4.00,4.76,6.72,7.71,8.45,8.03,8.79,8.79,8.80,7.23,6.13,4.53,4.31,2.22,2.01,1.05,0.66,0.13]
}, {
name: 'REP',
data: [0.00,0.09,0.09,0.95,1.21,1.67,2.24,3.22,3.70,6.24,6.43,6.62,8.01,8.42,8.97,8.48,7.45,7.68,8.64,7.37,2.53]
}],
[{
name: 'Dem',
data: [1.61,3.35,4.25,6.75,8.01,8.20,8.23,9.14,8.94,8.68,8.46,8.25,4.62,3.51,2.91,2.63,1.19,0.74,0.24,0.17,0.12]
},{
name: 'REP',
data: [0.21,0.38,0.68,1.62,1.55,2.55,3.99,4.65,4.31,5.78,6.28,6.79,8.47,9.01,8.61,8.34,7.16,6.50,6.10,4.78,2.25]
}],
[{
name: 'Dem',
data: [3.09,4.89,6.22,9.40,9.65,9.20,8.99,6.48,7.36,7.67,6.95,6.22,4.53,3.79,2.19,2.02,0.74,0.07,0.27,0.27,0.00]
}, {
name: 'REP',
data: [0.29,0.59,0.67,2.11,2.03,2.67,4.12,6.55,6.93,8.42,8.79,9.17,7.33,6.84,7.42,7.25,6.36,5.32,3.35,2.57,1.24]
}],
[{
name: 'Dem',
data: [6.00,5.24,5.11,7.66,9.25,8.25,8.00,8.09,8.12,7.05,6.59,6.12,4.25,4.07,2.30,1.49,0.98,0.80,0.42,0.16,0.06]
}, {
name: 'REP',
data: [0.00,0.13,0.13,0.48,0.97,2.10,2.73,3.14,3.64,5.04,5.30,5.56,6.87,6.75,8.03,9.33,11.01,10.49,7.61,6.02,4.68]
}],
[{
name: 'Dem',
data: [9.53,9.68,10.35,9.33,9.34,7.59,6.67,6.41,6.60,5.21,4.84,4.47,2.90,2.61,1.37,1.14,0.73,0.59,0.30,0.28,0.06]
}, {
name: 'REP',
data: [0.15,0.11,0.13,0.46,0.52,1.18,1.45,2.46,2.84,4.15,4.37,4.60,6.36,6.66,7.34,9.09,11.40,10.53,10.58,9.85,5.76]
}],
];
")
years <- c(1994, 1999, 2004, 2001, 2014)
# Transform the javascript data -------------------------------------------
party_data <- ctx$get("party_data")
map_df(1:length(party_data), function(i) {
x <- party_data[[i]]
names(x$data) <- x$name
dat <- as.data.frame(x$data)
bind_cols(dat, data_frame(x=-10:10, year=rep(years[i], nrow(dat))))
}) -> party_data
party_engaged_data <- ctx$get("party_engaged_data")
map_df(1:length(party_engaged_data), function(i) {
x <- party_engaged_data[[i]]
names(x$data) <- x$name
dat <- as.data.frame(x$data)
bind_cols(dat, data_frame(x=-10:10, year=rep(years[i], nrow(dat))))
}) -> party_engaged_data
# We need it in long form -------------------------------------------------
gather(party_data, party, pct, -x, -year) %>%
mutate(party=factor(party, levels=c("REP", "Dem"))) -> party_data_long
gather(party_engaged_data, party, pct, -x, -year) %>%
mutate(party=factor(party, levels=c("REP", "Dem"))) -> party_engaged_data_long
# Traditional frequency polygon plots -------------------------------------
gg <- ggplot()
gg <- gg + geom_ribbon(data=party_data_long,
aes(x=x, ymin=0, ymax=pct, fill=party, color=party), alpha=0.5)
gg <- gg + scale_x_continuous(expand=c(0,0), breaks=c(-8, 0, 8),
labels=c("Consistently\nliberal", "Mixed", "Consistently\nconservative"))
gg <- gg + scale_y_continuous(expand=c(0,0), limits=c(0, 12))
gg <- gg + scale_color_manual(name=NULL, values=c(Dem="#728ea2", REP="#cf6a5d"),
labels=c(Dem="Democrats", REP="Republicans"))
gg <- gg + guides(color="none", fill=guide_legend(override.aes=list(alpha=1)))
gg <- gg + scale_fill_manual(name=NULL, values=c(Dem="#728ea2", REP="#cf6a5d"),
labels=c(Dem="Democrats", REP="Republicans"))
gg <- gg + facet_wrap(~year, ncol=2, scales="free_x")
gg <- gg + labs(x=NULL, y=NULL,
title="Political Polarization, 1994-2014 (General Population)",
caption="Source: http://www.people-press.org/2014/06/12/section-1-growing-ideological-consistency/iframe/")
gg <- gg + theme_hrbrmstr_an(grid="")
gg <- gg + theme(panel.margin=margin(t=30, b=30, l=30, r=30))
gg <- gg + theme(legend.position=c(0.75, 0.1))
gg <- gg + theme(legend.direction="horizontal")
gg <- gg + theme(axis.text.y=element_blank())
gg
gg <- ggplot()
gg <- gg + geom_ribbon(data=party_engaged_data_long,
aes(x=x, ymin=0, ymax=pct, fill=party, color=party), alpha=0.5)
gg <- gg + scale_x_continuous(expand=c(0,0), breaks=c(-8, 0, 8),
labels=c("Consistently\nliberal", "Mixed", "Consistently\nconservative"))
gg <- gg + scale_y_continuous(expand=c(0,0), limits=c(0, 12))
gg <- gg + scale_color_manual(name=NULL, values=c(Dem="#728ea2", REP="#cf6a5d"),
labels=c(Dem="Democrats", REP="Republicans"))
gg <- gg + guides(color="none", fill=guide_legend(override.aes=list(alpha=1)))
gg <- gg + scale_fill_manual(name=NULL, values=c(Dem="#728ea2", REP="#cf6a5d"),
labels=c(Dem="Democrats", REP="Republicans"))
gg <- gg + facet_wrap(~year, ncol=2, scales="free_x")
gg <- gg + labs(x=NULL, y=NULL,
title="Political Polarization, 1994-2014 (Politically Active)",
caption="Source: http://www.people-press.org/2014/06/12/section-1-growing-ideological-consistency/iframe/")
gg <- gg + theme_hrbrmstr_an(grid="")
gg <- gg + theme(panel.margin=margin(t=30, b=30, l=30, r=30))
gg <- gg + theme(legend.position=c(0.75, 0.1))
gg <- gg + theme(legend.direction="horizontal")
gg <- gg + theme(axis.text.y=element_blank())
gg


It provides a similar effect to the Pew & Interworks visuals using alpha transparency to blend the point of polygon intersections. But I _really_ kinda like the way both Pew & Interworks did their visualizations without alpha blending yet still highlighting the intersected areas. We can do that in R as well with a bit more work by:
– grouping each data frame by year
– turning each set of points (Dem & Rep) to R polygons
– computing the intersection of those polygons
– turning that intersection back into a data frame
– adding this new polygon to the plots while also removing the alpha blend
Here’s what that looks like in code:
# Setup a function to do the polygon intersection -------------------------
polysect <- function(df) {
bind_rows(data_frame(x=-10, pct=0),
select(filter(df, party=="Dem"), x, pct),
data_frame(x=10, pct=0)) %>%
as.matrix() %>%
Polygon() %>%
list() %>%
Polygons(1) %>%
list() %>%
SpatialPolygons() -> dem
bind_rows(data_frame(x=-10, pct=0),
select(filter(df, party=="REP"), x, pct),
data_frame(x=10, pct=0)) %>%
as.matrix() %>%
Polygon() %>%
list() %>%
Polygons(1) %>%
list() %>%
SpatialPolygons() -> rep
inter <- gIntersection(dem, rep)
inter <- as.data.frame(inter@polygons[[1]]@Polygons[[1]]@coords)[c(-1, -25),]
inter <- mutate(inter, year=df$year[1])
inter
}
# Get the intersected area ------------------------------------------------
group_by(party_data_long, year) %>%
do(polysect(.)) -> general_sect
group_by(party_engaged_data_long, year) %>%
do(polysect(.)) -> engaged_sect
# Try the plots again -----------------------------------------------------
gg <- ggplot()
gg <- gg + geom_ribbon(data=party_data_long,
aes(x=x, ymin=0, ymax=pct, fill=party, color=party))
gg <- gg + geom_ribbon(data=general_sect, aes(x=x, ymin=0, ymax=y), color="#666979", fill="#666979")
gg <- gg + scale_x_continuous(expand=c(0,0), breaks=c(-8, 0, 8),
labels=c("Consistently\nliberal", "Mixed", "Consistently\nconservative"))
gg <- gg + scale_y_continuous(expand=c(0,0), limits=c(0, 12))
gg <- gg + scale_color_manual(name=NULL, values=c(Dem="#728ea2", REP="#cf6a5d"),
labels=c(Dem="Democrats", REP="Republicans"))
gg <- gg + guides(color="none", fill=guide_legend(override.aes=list(alpha=1)))
gg <- gg + scale_fill_manual(name=NULL, values=c(Dem="#728ea2", REP="#cf6a5d"),
labels=c(Dem="Democrats", REP="Republicans"))
gg <- gg + facet_wrap(~year, ncol=2, scales="free_x")
gg <- gg + labs(x=NULL, y=NULL,
title="Political Polarization, 1994-2014 (General Population)",
caption="Source: http://www.people-press.org/2014/06/12/section-1-growing-ideological-consistency/iframe/")
gg <- gg + theme_hrbrmstr_an(grid="")
gg <- gg + theme(panel.margin=margin(t=30, b=30, l=30, r=30))
gg <- gg + theme(legend.position=c(0.75, 0.1))
gg <- gg + theme(legend.direction="horizontal")
gg <- gg + theme(axis.text.y=element_blank())
gg
gg <- ggplot()
gg <- gg + geom_ribbon(data=party_engaged_data_long,
aes(x=x, ymin=0, ymax=pct, fill=party, color=party))
gg <- gg + geom_ribbon(data=engaged_sect, aes(x=x, ymin=0, ymax=y), color="#666979", fill="#666979")
gg <- gg + scale_x_continuous(expand=c(0,0), breaks=c(-8, 0, 8),
labels=c("Consistently\nliberal", "Mixed", "Consistently\nconservative"))
gg <- gg + scale_y_continuous(expand=c(0,0), limits=c(0, 12))
gg <- gg + scale_color_manual(name=NULL, values=c(Dem="#728ea2", REP="#cf6a5d"),
labels=c(Dem="Democrats", REP="Republicans"))
gg <- gg + guides(color="none", fill=guide_legend(override.aes=list(alpha=1)))
gg <- gg + scale_fill_manual(name=NULL, values=c(Dem="#728ea2", REP="#cf6a5d"),
labels=c(Dem="Democrats", REP="Republicans"))
gg <- gg + facet_wrap(~year, ncol=2, scales="free_x")
gg <- gg + labs(x=NULL, y=NULL,
title="Political Polarization, 1994-2014 (Politically Active)",
caption="Source: http://www.people-press.org/2014/06/12/section-1-growing-ideological-consistency/iframe/")
gg <- gg + theme_hrbrmstr_an(grid="")
gg <- gg + theme(panel.margin=margin(t=30, b=30, l=30, r=30))
gg <- gg + theme(legend.position=c(0.75, 0.1))
gg <- gg + theme(legend.direction="horizontal")
gg <- gg + theme(axis.text.y=element_blank())
gg


Without much extra effort/work we now have what I believe to be a more striking set of visuals. (And, I should probably makes a `points_to_spatial_polys()` convenience function.)
You’ll find the “overall” group data as well as the party median values in [the Pew HTML source code](view-source:http://www.people-press.org/2014/06/12/section-1-growing-ideological-consistency/iframe/) if you want to try to fully replicate their visualizations.
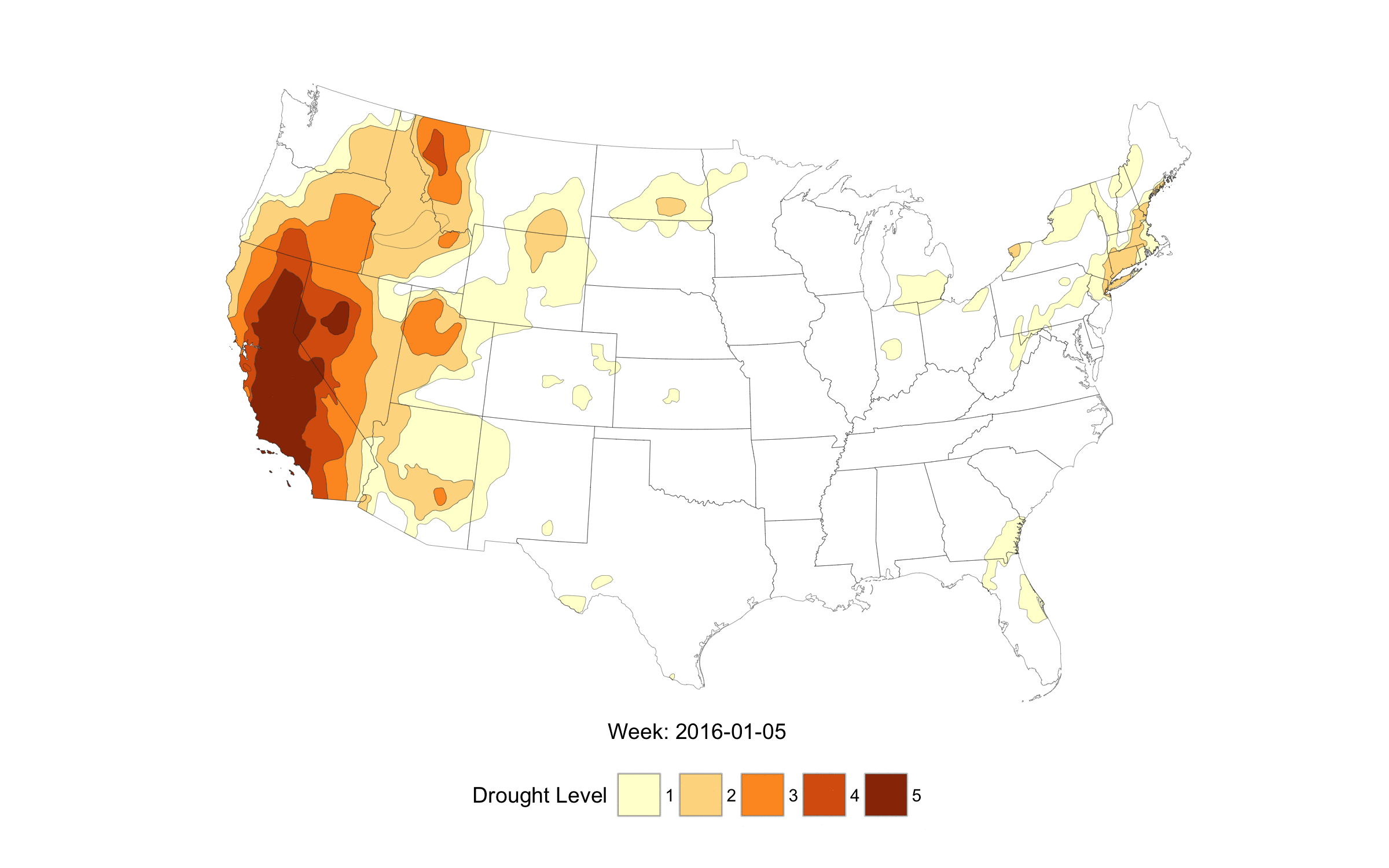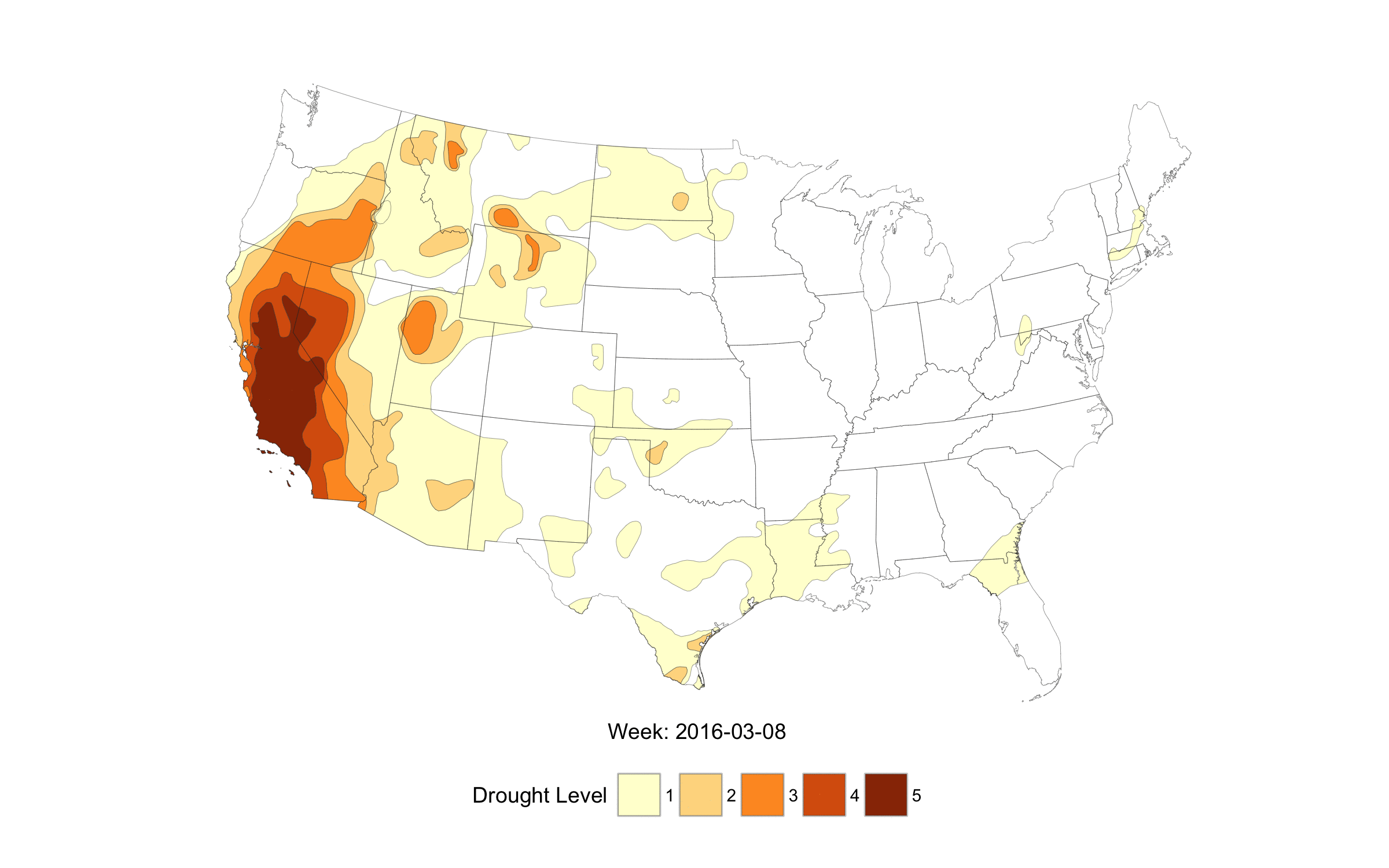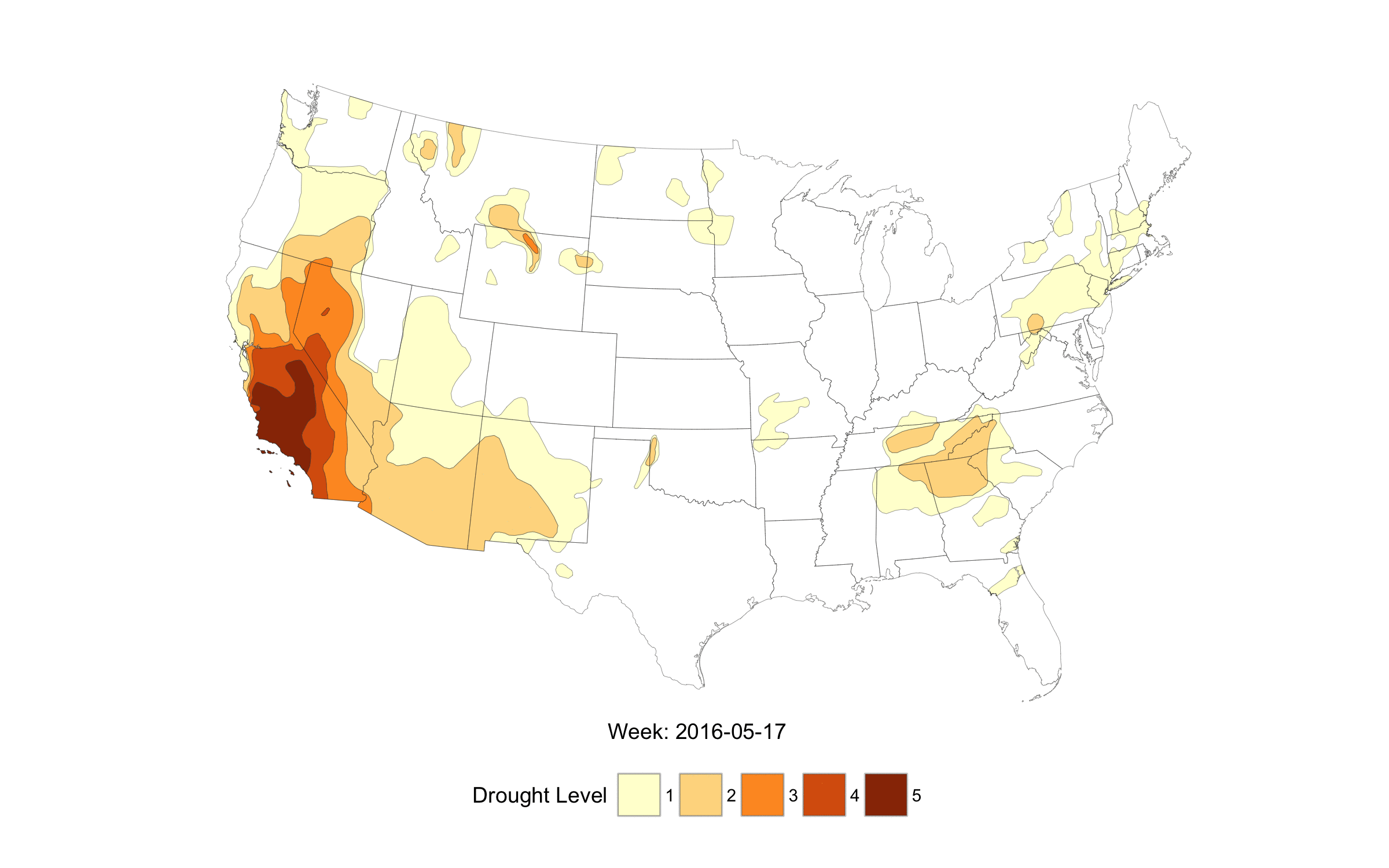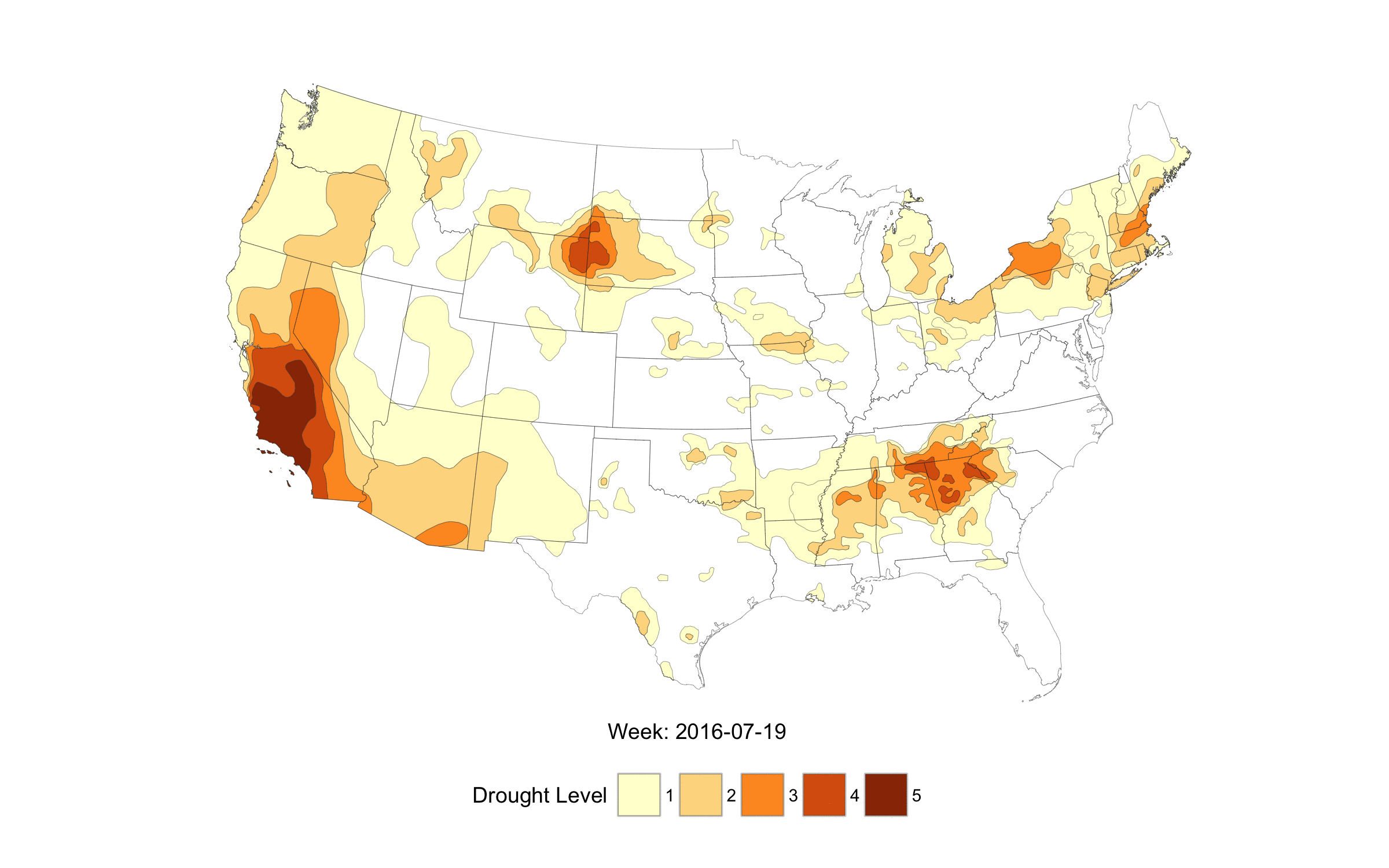






When life imitates Fullmetal Alchemist
The NIH is [moving forward](http://www.npr.org/sections/health-shots/2016/08/04/488387729/nih-plans-to-lift-ban-on-research-funds-for-part-human-part-animal-embryos) with plans to financially support & encourage human-animal chimera research.
You can find more info over at the [NIH blog](http://osp.od.nih.gov/under-the-poliscope/2016/08/next-steps-research-using-animal-embryos-containing-human-cells).
Chimera’s have been a longstanding subject of science-fiction/fantasy and many authors have visited it to help inform the ethics debate. A fairly recent exploration of this has been through the [Fullmetal Alchemist](http://www.fullmetalalchemist.com/) anime/manga series. TLDR: it doesn’t go so well, even in animal-animal chimera hybrids.
Yes, that’s fiction and the current NIH proposals are nowhere near as audacious as what’s described in the FMA series. But, surprisingly, you can’t find a large number vocal critics of human-animal chimera research since the modern “open scientific community” is actually pretty harshly judgmental of anyone that tries to limit or challenge “science” in any way (since it’s their religion, as everyone believes in something whether they claim to or not). Open, logical and—more importantly—effective criticisms against purported “progress” are often career-limiting moves. All this at a time in history when the current generation of scientists seems to be excelling at ignoring the potential for unintended consequences of their works.
Folks can (and should) [add their comments](http://grants.nih.gov/grants/rfi/rfi.cfm?ID=57) either for or against this proposal. Not commenting means you agree with the NIH plans and support your U.S. tax dollars & government resources going to support this research; it also means you are on the hook when this eventually goes horribly, horribly wrong.
For those commenting to show their _lack_ of support, I augmented a statement from an [interview wtih Dr. Stuart Newman](http://www.beliefnet.com/news/science-religion/2005/05/the-peril-and-promise-of-mix-and-match-biotech.aspx), who is a vocal detractor of human-animal chimeras (so much so that he tried to prevent it through the USPTO process, which eventually failed) for my submission:
As the translation from the FMA series states: _”Humans have a limitless desire to use their knowledge in real life…the desire to see what you can do with the power that is given to them…the desire to understand all the secrets in this world and experiment with them.”_ My reasoning and my faith suggest that there are definitely doors that should remain closed.