It’s been a while since I’ve posted anything R-related and, while this one will be brief, it may be of use to some R folks who have taken the leap into Big Sur and/or Apple Silicon. Stay to the end for an early Christmas 🎁!
Big Sur Report
As #rstats + #macOS Twitter-folks know (before my permanent read hiatus began), I’ve been on Big Sur since the very first developer betas were out. I’m even on the latest beta (11.1b1) as I type this post.
Apple picked away at many nits that they introduced with Big Sur, but it’s still more of a “Vista” release than Catalina was given all the clicks involved with installing new software. However, despite making Simon’s life a bit more difficult (re: notarization of R binaries) I’m happy to report that R 4.0.3 and the latest RStudio 1.4 daily releases work great on Big Sur. To be on the safe side, I highly recommend putting the R command-line binaries and RStudio and R-GUI .apps into both “Developer Tools” and “Full Disk Access” in the Security & Privacy preferences pane. While not completely necessary, it may save some debugging (or clicks of “OK”) down the road.
The Xcode command-line tools are finally in a stable state and can be used instead of the massive Xcode.app installation. This was problematic for a few months, but Apple has been pretty consistent keeping it version-stable with Xcode-proper.
Homebrew is pretty much feature complete on Big Sur (for Intel-architecture Macs) and I’ve run everything from the {tidyverse} to {sf}-verse, ODBC/JDBC and more. Zero issues have come up, and with the pending public release (in a few weeks) of 11.1, I can safely say you should be fine moving your R analyses and workflows to Big Sur.
Apple Silicon Report
Along with all the other madness, 2020 has resurrected the “Processor Wars” by making their own ARM 64-bit chips (the M1 series). The Mac mini flavor is fast, but I suspect some of the “feel” of that speed comes from the faster SSDs and beefed up I/O plane to get to those SSDs. These newly released Mac models require Big Sur, so if you’re not prepared to deal with that, you should hold off on a hardware upgrade.
Another big reason to hold off on a hardware upgrade is that the current M1 chips cannot handle more than 16GB of RAM. I do most of the workloads requiring memory heavy-lifting on a 128GB RAM, 8-core Ubuntu 20 server, so what would have been a deal breaker in the past is not much of one today. Still, R folks who have gotten used to 32GB+ of RAM on laptops/desktops will absolutely need to wait for the next-gen chips.
Most current macOS software — including R and RStudio — is going to run in the Rosetta 2 “translation environment”. I’ve not experienced the 20+ seconds of startup time that others have reported, but RStudio 1.4 did take noticeably (but not painfully) longer on the first run than it has on subsequent ones. Given how complex the RStudio app is (chromium engine, Qt, C++, Java, oh my!) I was concerned it might not work at all, but it does indeed work with absolutely no problems. Even the ODBC drivers (Athena, Drill, Postgres) I need to use in daily work all work great with R/RStudio.
This means things can only get even better (i.e. faster) as all these components are built with “Universal” processor support.
Homebrew can work, but it requires the use of the arch command to ensure everything is running the Rosetta 2 translation environment and nothing I’ve installed (from source) has required anything from Homebrew. Do not let that last sentence lull you into a false sense of excitement. So far, I’ve tested:
- core {tidyverse}
- {DBI}, {odbc}, {RJDBC}, and (hence) {dbplyr}
- partially extended {ggplot2}-verse
- {httr}, {rvest}, {xml2}
- {V8}
- a bunch of self-contained, C[++]-backed or just base R-backed stats packages
and they all work fine.
I have installed separate (non-Universal) binaries of fd, ripgrep, bat plus a few others, and almost have a working Rust toolchain up (both Rust and Go are very close to stable on Apple’s M1 architecture).
If there are specific packages/package ecosystems you’d like tested or benchmarked, drop a note in the comments. I’ll likely bee adding more field report updates over the coming weeks as I experiment with additional components.
Now, if you are a macOS R user, you already know — thanks to Tomas and Simon — that we are in wait mode for a working Fortran compiler before we will see a Universal build of R. The good news is that things are working fine under Rosetta 2 (so far).
RStudio Update
If there is any way you can use RStudio Desktop + Server Pro, I would heartily recommend that you do so. The remote R session in-app access capabilities are dreamy and addictive as all get-out.
I also (finally) made a (very stupid simple) PR into the dailies so that RStudio will be counted as a “Developer Tool” for Screen Time accounting.
RSwitch Update
🎁-time! As incentive to try Big Sur and/or Apple Silicon, I started work on version 2 of RSwitch which you can grab from — https://rud.is/rswitch/releases/RSwitch-2.0.0b.app.zip. For those new to RSwitch, it is a modern alternative to the legacy RSwitch that enabled easy switching of R versions on macOS.
The new version requires Big Sur as its based on Apple’s new “SwiftUI” and takes advantage of some components that Apple has not seen fit to make backwards compatible. The current version has fewer features than the 1.x series, but I’m still working out what should and should not be included in the app. Drop notes in the comments with feature requests (source will be up after 🦃 Day).
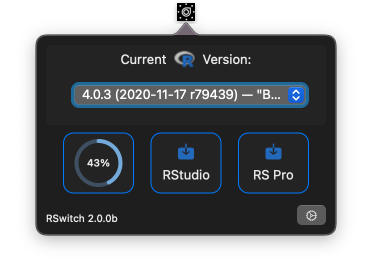
The biggest change is that the app is not just a menu but an popup window:
Each of those tappable download regions presents a cancellable download progress bar (all three can run at the same time), and any downloaded disk images will be auto-mounted. That third tappable download region is for downloading RStudio Pro dailies. You can get notifications for both (or neither) Community and Pro versions:
The R version switchers also provides more info about the installed versions:
(And, yes, the r79439 is what’s in Rversion.h of each of those installations. I kinda want to know why, now.)
The interface is very likely going to change dramatically, but the app is a Universal binary, so will work on M1 and Intel Big Sur Macs.
NOTE: it’s probably a good idea to put this into “Full Disk Access” in the Security & Privacy preferences pane, but you should likely wait until I post the source so you can either build it yourself or validate that it’s only doing what it says on the tin for yourself (the app is benign but you should be wary of any app you put on your Macs these days).
WARNING: For now it’s a Dark Mode only app so if you need it to be non-hideous in Light Mode, wait until next week as I add in support for both modes.
FIN
If you’re running into issues, the macOS R mailing list is probably a better place to post issues with R and BigSur/Apple Silicon, but feel free to drop a comment if you are having something you want me to test/take a stab at, and/or change/add to RSwitch.
(The RSwitch 2.0.0b release yesterday had a bug that I think has been remedied in the current downloadable version.)